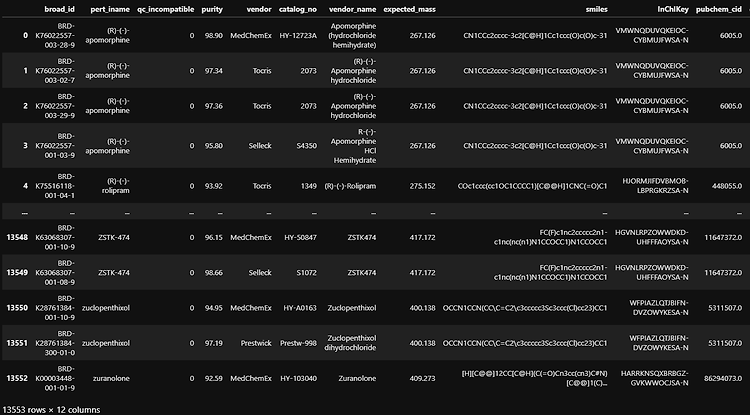
Pandas concat vs append vs join vs merge Concat gives the flexibility to join based on the axis( all rows or all columns) Append is the specific case(axis=0, join='outer') of concat Join is based on the indexes (set by set_index) on how variable =['left','right','inner','couter'] Merge is based on any particular column each of the two dataframes, this columns are variables on like 'left_on', 'ri..